Research InterestsResearch in the Seaman Lab focuses on understanding the molecular basis of phenotypic adaptation in primates, with an emphasis on recent evolution in humans and the great apes. Using a variety of wet lab techniques and computational approaches, we seek to uncover what changes at what genes are responsible for the anatomical and physiological differences among primate species. Our lab is also interested in patterns of genome evolution, particularly the biology of numts and evolution via gene duplication. Details on specific projects are given below. If you are interested in joining our group as an undergraduate or graduate student please contact Dr. Seaman directly. |
||
|
Molecular evolution of reproductive proteinsMany adaptations related to reproduction have occurred during the evolution of humans and our closest relatives, the apes. Among these are the "invasive placenta", a longer gestation, and larger newborns. In addition to these general trends of hominoids, species-specific adaptations to unique mating systems and social behaviors include variations in testes size, characteristics of semen, and secondary sexual characters. Previous research in our lab has examined the evolution of coding sequences of several male reproductive genes, uncovering episodes of rapid adaptive evolution at the molecular level, along with loss of function of several genes (Jensen-Seaman & Li 2003; Carnahan & Jensen-Seaman 2008).Current research in this area is taking a more in-depth and multifaceted approach to understanding the molecular evolution of proteins found in ejaculated semen. We are examining the mode and tempo of the evolution of both protein-coding and promoter sequences of the genes that code for such proteins. We are using in vitro assays to quantify the differences among hominoid species in the ability of their promoters to drive transcription at male reproductive genes. Finally, we are using proteomic approaches to characterize the protein constituents in the seminal plasma of several primate species. The results are being interpreted in light of natural and sexual selection, and the unique selective pressures seen among hominoid species. Recent publications from this project include Carnahan-Craig & Jensen-Seaman (2014). This work is supported by the National Science Foundation, Division of Physical Anthropology, and the Wenner-Gren Foundation for Anthropological Research. |
|
|
 |
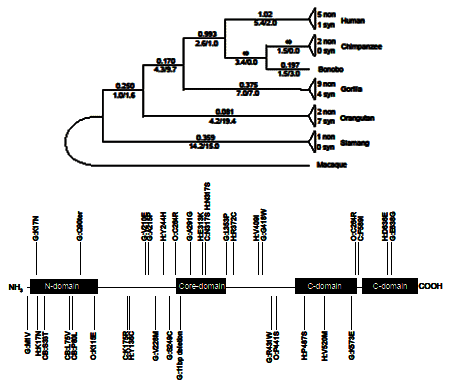
Evolution of the relaxin genesThe relaxin peptide hormones are essential for several aspects of pregnancy and birth. The genomic
cluster containing the relaxin genes, and other related genes of a larger superfamily, has experienced gene duplications, gene conversion,
and pseudogenization during recent hominoid evolution. In addition, the sequence of the coding regions and the promoter regions show
substantial variation both within and among species. With this background of genomic evolution, we are investigating the evolutionary
history of the region and the functional consequences of genetic variation at the relaxin genes.
|
|
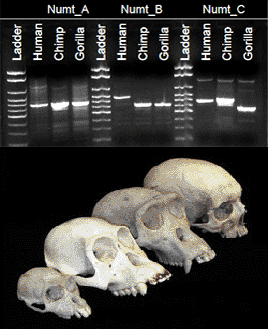
Numt evolution in hominoid primatesMy lab has had a long-standing interest in the integration of mitochondrial DNA into nuclear DNA (numts), particularly in gorillas
(Jensen-Seaman et al. 2004). Although
mitochondrial DNA is often used in population genetics and phylogenetics, results can be confounded by inadvertent inclusion of numts.
We are currently collaborating with the lab of Dr. Nicola Anthony
of the University of New Orleans
to study the rates and patterns of numt integration in humans, chimpanzees, and gorillas; their relevance for phylogeographic studies of
African apes; and the potential use of polymorphic numt insertions as molecular markers in these species. For results, see our published papers
from this project (Jensen-Seaman et al. 2009;
Das et al. 2014 ;Soto-Calderón et al. 2014).
|
|
|

|
Distribution of a novel metal-binding motifOur lab maintains a collaboration with Dr. Charles Dameron of Saint Francis University, who is interested in metal binding proteins and metalloregulation. Our contribution includes the bioinformatic analysis of complete genome sequences of prokaryotes and eukaryotes to determine the distribution and evolutionary history of genes and gene families containing a recently described metal-binding and regulatory protein motif.
|
|
FundingWork in my laboratory has been funded by the following organizations: |
||